Class I Immunogenicity - Tutorial
This tool uses amino acid properties as well as their position within the peptide to predict the immunogenicity of a peptide MHC (pMHC) complex.
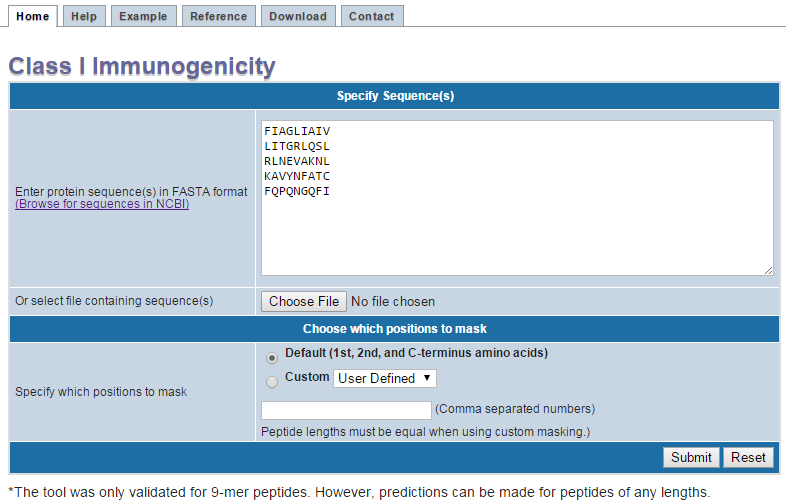
1: Enter peptide sequences.
Enter peptide sequences separated by a new line into the text field. Ideally peptides should be the same length, presented on the same HLA class I molecule,
and 9-mers. This method was
validated only on 9mers presented on HLA class I molecules. When longer peptides are provided, the extra amino acids will be evaluated as if they are inserted after
position 5, and as if they have the same properties as an amino acid at position 5 in terms of weighting and masking.
validated only on 9mers presented on HLA class I molecules. When longer peptides are provided, the extra amino acids will be evaluated as if they are inserted after
position 5, and as if they have the same properties as an amino acid at position 5 in terms of weighting and masking.
2: Upload peptide file.
As alternative to manually entering peptide sequences, a user can upload a standard text file with sequences. Each sequence should be entered on its own line.
3: Choose masking option.
This will mask designated peptides from the immunogenicity score. The masked positions are dependent on the HLA molecule on which the peptide was presented.
As a default, the first,
second, and C-terminal amino acid are masked. Additional anchor positions are used for some HLA molecules. For most common HLA class I molecules the default masking scheme can be
selected. A custom mask scheme can be entered by inputting comma separated numbers. Note that the custom masking option will only work when peptides of the same length are provided.
second, and C-terminal amino acid are masked. Additional anchor positions are used for some HLA molecules. For most common HLA class I molecules the default masking scheme can be
selected. A custom mask scheme can be entered by inputting comma separated numbers. Note that the custom masking option will only work when peptides of the same length are provided.
4: Submit or reset form.
Submit your peptides for calculation or reset the current form.
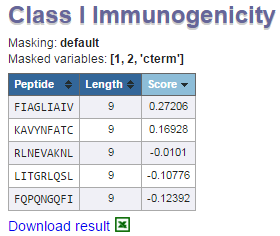
5: Masking of results.
Displays the masking options chosen.
6: Masking variables chosen
Displaying which amino acids were masked in the calculations.
7: Table of results.
Each row in this table corresponds to prediction for an individual peptide. The table consists of peptide, length and score columns. By default prediction results
are sorted by descending score values. However, the table can also be sorted by clicking on individual column headers. The higher score indicates a greater
probability of eliciting an immune response.
8: Generate Text Results.
Generates text file of peptide calculations within the web browser.