Methods for modeling and docking of antibody and protein 3D structures
This page provides information on methods for modeling and docking of antibody and protein 3D structures. The methods are available as web servers and are hosted
outside the IEDB except one method, LYRA.
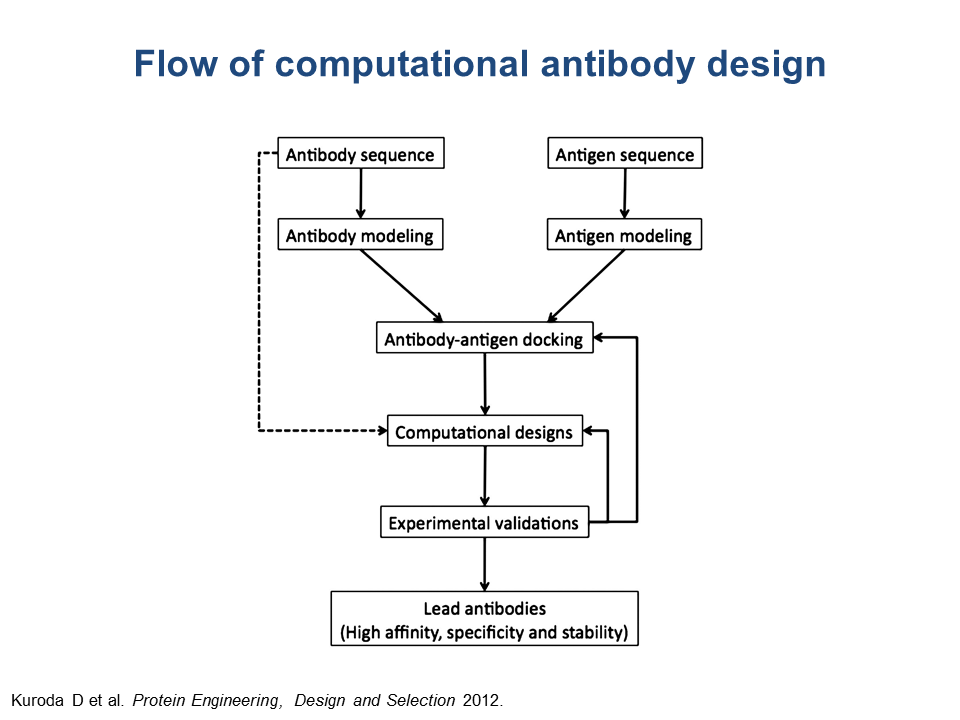
One of the approaches at B-cell epitope prediction involves antibody-antigen docking, or 3D modeling of an antibody-antigen complex, for known 3D structures of both antibody and antigen. When the structures of either antibody or protein or both are unknown (that is, not available in PDB) but sequences are known, the user can attempt to obtain the 3D structural models of the antibody and the antigen, following the chart shown below and using the described web servers.
Web servers for modeling antibody 3D structure(operational as of November 2014)
These methods require the user to submit sequences of both light and heavy chains of the antibody. The output of either method is the 3D structure that can be downloaded as a text file in the PDB format.
For example, using LYRA, one can model the structure of Tocilizumab, the antibody used as an immunosuppressive drug for treatment of rheumatoid arthritis and systemic juvenile idiopathic arthritis. The sequences of the heavy and light chains of this and many other commercial antibodies can be found at http://imgt.org/mAb-DB/query. Here are the obtained sequences of Tocilizumab that can be paste in LYRA:
Web servers for modeling protein 3D structure and databases of protein models (operational as of November 2014)
Web servers for modeling 3D structure of antibody-protein complexes (operational as of November 2014) – assume antibody is a receptor and protein antigen is a ligand
Docking methods output multiple possible predicted 3D structures of antibody-antigen complexes that can be downloaded as pdb-formatted files. The docking approach is unable to predict antibody binding sites de novo with high accuracy; however, if the binding site is at least somewhat defined (for example, through truncation experiments), it might be possible to arrive at one or a few most likely predicted conformations that can be tested further experimentally, for example, using the site-directed mutagenesis. Please see the IEDB workshop 2014 slides (download 5D-PIGS2014Day2.pdf) for more information.
One of the approaches at B-cell epitope prediction involves antibody-antigen docking, or 3D modeling of an antibody-antigen complex, for known 3D structures of both antibody and antigen. When the structures of either antibody or protein or both are unknown (that is, not available in PDB) but sequences are known, the user can attempt to obtain the 3D structural models of the antibody and the antigen, following the chart shown below and using the described web servers.

Web servers for modeling antibody 3D structure(operational as of November 2014)
- LYRA (http://tools.iedb.org/lyra/)
- RosettaAntibody (http://antibody.graylab.jhu.edu/)
These methods require the user to submit sequences of both light and heavy chains of the antibody. The output of either method is the 3D structure that can be downloaded as a text file in the PDB format.
For example, using LYRA, one can model the structure of Tocilizumab, the antibody used as an immunosuppressive drug for treatment of rheumatoid arthritis and systemic juvenile idiopathic arthritis. The sequences of the heavy and light chains of this and many other commercial antibodies can be found at http://imgt.org/mAb-DB/query. Here are the obtained sequences of Tocilizumab that can be paste in LYRA:
>Heavy chain QVQLQESGPGLVRPSQTLSLTCTVSGYSITSDHAWSWVRQPPGRGLEWIGYISYSGITTYNPSLKSR VTMLRDTSKNQFSLRLSSVTAADTAVYYCARSLARTTAMDYWGQGSLVTVSSASTKGPSVFPLAPSS KSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYI CNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV SHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIE KTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDS DGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK >Light chain DIQMTQSPSSLSASVGDRVTITCRASQDISSYLNWYQQKPGKAPKLLIYYTSRLHSGVPSRFSGSGS GTDFTFTISSLQPEDIATYYCQQGNTLPYTFGQGTKVEIKRTVAAPSVFIFPPSDEQLKSGTASVVC LLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
Web servers for modeling protein 3D structure and databases of protein models (operational as of November 2014)
- Protein Model Portal (http://www.proteinmodelportal.org/)
- SWISS-MODEL Repository (SMR) - a database of annotated 3D protein structure models (http://swissmodel.expasy.org/repository/)
- ModBase – a database of comparative protein structure models (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi)
Web servers for modeling 3D structure of antibody-protein complexes (operational as of November 2014) – assume antibody is a receptor and protein antigen is a ligand
- Cluspro 2.0 (http://cluspro.bu.edu/login.php ) -- make sure to select “Advanced Options” -> “Antibody Mode” and select check boxes “Use Antibody Mode” and “Automatically Mask non-CDR regions”
- PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock/) – make sure to select “Antibody-antigen” for the “Complex Type”
Docking methods output multiple possible predicted 3D structures of antibody-antigen complexes that can be downloaded as pdb-formatted files. The docking approach is unable to predict antibody binding sites de novo with high accuracy; however, if the binding site is at least somewhat defined (for example, through truncation experiments), it might be possible to arrive at one or a few most likely predicted conformations that can be tested further experimentally, for example, using the site-directed mutagenesis. Please see the IEDB workshop 2014 slides (download 5D-PIGS2014Day2.pdf) for more information.